CMS Data Analysis School Pre-Exercises - Fourth Set
Overview
Teaching: 0 min
Exercises: 60 minQuestions
How do we analyze an EDM ROOT file using an EDAnalyzer?
How do we analyze an EDM ROOT file using an FWLite executable?
How do we use ROOT/RooFit to fit a function to a histogram?
Objectives
Learn how to use an EDAnalyzer
Learn how to use FWLite
Understand a variety of methods for performing a fit to a histogram
Introduction
In this set of exercises, we will analyze the MiniAOD file that was made in the third set of exercise. You must have this skimmed MiniAOD stored locally (in your eos user space) in order to access them. We will use several different workflows for analyzing the MiniAOD, namely an EDAnalyzer, a FWLite executable, a FWLite Macro, and a FWLite PyROOT script. We will basically re-make the Z peak and few other histograms and store them in an output root file. In the exercise in the end we will try to fit with a Gaussian, Breit-Wigner function, etc.
Warning
To perform this set of exercises, a CERN computing account, Grid Certificate, and CMS VO membership are required. You should already have these things, but if not, follow these instructions from the setup instructions.
Objective
Please post your answers to the questions in the Google form fourth set.
Exercise 15 - Analyzing MiniAOD with an EDAnalyzer
In this exercise we will analyze the skimmed MiniAODs created in the third set of exercises using an EDAnalyzer. In these skimmed MiniAODs, if you recall, we saved only the muons and electrons. So do not look for jets, photons, or other objects as they were simply not saved. We will use a python config file and an EDAnalyzer ( a .cc file) to make a Z mass peak. You can find an example list of files below, but please first try using the files you created.
Example file list
root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_1.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_10.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_11.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_12.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_13.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_14.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_15.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_16.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_17.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_18.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_19.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_2.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_20.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_21.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_22.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_23.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_24.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_25.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_26.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_27.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_28.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_29.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_3.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_30.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_31.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_4.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_5.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_6.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_7.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_8.root root://cmseos.fnal.gov//eos/uscms/store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/slimMiniAOD_data_MuEle_9.root
First we will add the PhysicsTools/PatExamples package as follows to <YOURWORKINGAREA>/CMSSW_10_6_18/src. The PatExamples package has lot of examples for a user to try. However, we will add our own code and config file to it and then compile. To add this package, do this:
cd $CMSSW_BASE/src/
git cms-addpkg PhysicsTools/PatExamples
Note
We are assuming that you’ve already checked out a CMSSW_10_6_18 release and have performed the
cmsenvsetup command.
In this package, you will find the python configuration file $CMSSW_BASE/src/PhysicsTools/PatExamples/test/analyzePatBasics_cfg.py. You will also see the EDAnalyzer in $CMSSW_BASE/src/PhysicsTools/PatExamples/plugins/PatBasicAnalyzer.cc.
Next, create the following two files (download/save): $CMSSW_BASE/src/PhysicsTools/PatExamples/src/MyZPeakAnalyzer.cc and $CMSSW_BASE/src/MyZPeak_cfg.py.
Hint
A quick way to do this on Linux, or any machine with
wget, is by using the following commands:wget https://cern-cms-das-2023.github.io/cms-das-pre-exercises/code/MyZPeakAnalyzer-CMSSW_10_6_18.cc -O $CMSSW_BASE/src/PhysicsTools/PatExamples/src/MyZPeakAnalyzer.cc wget https://cern-cms-das-2023.github.io/cms-das-pre-exercises/code/MyZPeak_cfg.py -O $CMSSW_BASE/src/MyZPeak_cfg.py
Then we will compile the code that you just saved by doing:
cd $CMSSW_BASE/src/
scram b
The compilation should print many lines of text to your terminal. Among those lines you should see a line like the one below. If you can’t find a similar line, then the code you just added is not compiled.
>> Compiling <$CMSSW_BASE>/src/PhysicsTools/PatExamples/src/MyZPeakAnalyzer.cc
After successful compilation, you must run the config file as follows:
cmsRun MyZPeak_cfg.py
Successful running of the above config file will produce an output file myZPeakCRAB.root. The output file myZPeakCRAB.root has several histograms, besides the Z peak, called mumuMass, like muonMult, muonEta, muonPhi, muonPt and similarly for electrons.
Note
In the case above, the file
MyZPeak_cfg.py will read from arearoot://cmseos.fnal.gov//store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/. You should have a similar location from where you can read your CRAB outputROOTfiles. You can edit theMyZPeak_cfg.pyfile to use the MiniAOD files you made in Exercise 13 by replacing the location of the input files to the path of file you generated. From my side, the files are stored in:'/eos/user/v/vmilosev/DoubleMuon/crab_CMSDAS_Data_analysis_test0/230421_160319/0000/slimMiniAOD_data_MuEle_1.root'
Question 15
What is the number of entries in the
mumuMassplot if you just used the first input file, probably namedslimMiniAOD_data_MuEle_1.root?
Exercise 16 - Analyzing MiniAOD with an FWLite executable
In this exercise we will make the same ROOT file, myZPeakCRAB.root, as in Exercise 15, but we call it myZPeakCRAB_fwlite.root so that you do not end up overwriting the file previously made in Exercise 15.
First, check out the following two packages by doing:
cd $CMSSW_BASE/src/
git cms-addpkg PhysicsTools/FWLite
git cms-addpkg PhysicsTools/UtilAlgos
Next, replace the existing $CMSSW_BASE/src/PhysicsTools/FWLite/bin/FWLiteWithPythonConfig.cc with this FWLiteWithPythonConfig.cc. You are simply updating an existing analyzer. Then, create the file $CMSSW_BASE/src/parameters.py.
Hint
You can easily download the needed files by running the following commands:
wget https://cern-cms-das-2023.github.io/cms-das-pre-exercises/code/FWLiteWithPythonConfig.cc -O $CMSSW_BASE/src/PhysicsTools/FWLite/bin/FWLiteWithPythonConfig.cc wget https://cern-cms-das-2023.github.io/cms-das-pre-exercises/code/parameters.py -O $CMSSW_BASE/src/parameters.py
Note
In case you have completed Exercise Set 3 successfully, put the names and path of the
ROOTfiles that you made yourself via submitting CRAB job, instead of those currently inparameters.py.
parameters.pywill read from arearoot://cmseos.fnal.gov//store/user/cmsdas/2023/pre_exercises/Set4/Input/DoubleMuon/. You should have a similar location from where you can read your CRAB outputROOTfiles. You can edit theparameters.pyfile to use the MiniAOD files you made in Exercise 13 by replacing the location of the input files. From my side, the files are stored in:'/eos/user/v/vmilosev/DoubleMuon/crab_CMSDAS_Data_analysis_test0/230421_160319/0000/slimMiniAOD_data_MuEle_1.root'
Then we will compile the code that you just saved by doing:
cd $CMSSW_BASE/src/
scram b -j 4
You should see among the output a line like the one below. If not, it is probable that you haven’t compiled the code on which we are working.
>> Compiling /your_path/YOURWORKINGAREA/CMSSW_10_6_18/src/PhysicsTools/FWLite/bin/FWLiteWithPythonConfig.cc
After successful compilation, you must run the config file as follows:
cd $CMSSW_BASE/src/
cmsenv
FWLiteWithPythonConfig parameters.py
Note
Take note of the extra
cmsenvis to ensure the changes to files in thebinsubdirectory are picked up in your path.
Warning
You might get a segfault when running this exercise. Just ignore it; the output
ROOTfile will still be created and be readable.
Note
Take a look at how the parameters defined in
parameters.pyget input to the executable codeFWLiteWithPythonConfig.cc.
A successful running of the FWLite executable, FWLiteWithPythonConfig, results in an output file called myZPeakCRAB_fwlite.root.
The output ROOT file myZPeakCRAB_fwlite.root is a bit different from myZPeakCRAB.root made in Exercise 15 since we did not make any of the electron histograms. The histograms do have the mumuMass, besides, muonEta, muonPhi, and muonPt.
Question 16
What is the number in entries in the
mumuMassobtained in Exercise 16, again using only the first input file.?
Exercise 17 - Fitting the Z mass peak
The main intention of fitting the Z mass peak is to show how to fit a distribution. To do this exercise, you will need the ROOT files that you made in Exercise 15 and Exercise 16. Make sure you have the ROOT file $CMSSW_BASE/src/myZPeakCRAB.root ( Exercise 15) or myZPeakCRAB_fwlite.root (Exercise 16). If you have not managed to create at least one of these ROOT files, you can get them from the following locations:
File list
/afs/cern.ch/cms/Tutorials/TWIKI_DATA/CMSDataAnaSch/myZPeakCRAB.root # lxplus or Bari root://cmseos.fnal.gov//store/user/cmsdas/2023/pre_exercises/Set4/Output/myZPeakCRAB.root # cmslpc root://cmseos.fnal.gov//store/user/cmsdas/2023/pre_exercises/Set4/Output/myZPeakCRAB_fwlite.root # cmslpc
This will allow you to continue with Exercise 17. For this exercise, we will use the ROOT file myZPeakCRAB.root. Alternatively, you can use the file myZPeakCRAB_fwlite.root, but just make sure to have the right name of the ROOT file. The most important factor is that both of these files have the histogram mumuMass.
We also ask that you create a rootlogon.C file in the $CMSSW_BASE/src/ directory. We will reference this version as opposed to anyone’s personalized rootlogon file. This sets up the libraries needed to complete this exercise.
The different distribution that we would fit to the Z mass peak are:
- Gaussian
- Relativistic Breit-Wigner
- Convolution of relativistic Breit-Wigner plus interference term with a Gaussian
Some general remarks about fitting a Z peak
To fit a generator-level Z peak, a Breit-Wigner fit makes sense. However, reconstructed-level Z peaks have many detector resolutions that smear the Z mass peak. If the detector resolution is relatively poor, then it is usually good enough to fit a Gaussian (since the gaussian detector resolution will overwhelm the inherent Briet-Wigner shape of the peak). If the detector resolution is fairly good, then another option is to fit a Breit-Wigner (for the inherent shape) convolved with a Gaussian (to describe the detector effects). This is in the “no-background” case. If you have backgrounds in your sample (Drell-Yan, cosmics, etc…), and you want to do the fit over a large mass range, then another function needs to be included to take care of this; an exponential is commonly used.
Fitting a Gaussian
There are several options to fit a Gaussian
Using the inbuilt Gaussian in ROOT
Open ROOT as follows:
root -l
Then execute the following commands:
TFile f("myZPeakCRAB.root");
f.cd("analyzeBasicPat");
gStyle->SetOptFit(111111);
mumuMass->Fit("gaus");
This will pop up the following histogram. Save this histogram as pdf, ps, or eps file using the menu of the histogram window. As you can see we should fit a sub-range as this fit is not a good fit. In the next part of this exercise, we will fit a sub-range of the mumuMass distribution, but for this we will use a ROOT macro as using inbuilt ROOT functions have very minimal usage. For more complex or useful fitting functions, one has to use a macro.
For now, we can improve the fit description of the Z resonance by limiting our fit range:
TFile f("myZPeakCRAB.root");
f.cd("analyzeBasicPat");
gStyle->SetOptFit(111111);
g1 = new TF1("m1","gaus",85,95);
mumuMass->Fit(g1,"R");
One should obtain a similar histogram as:
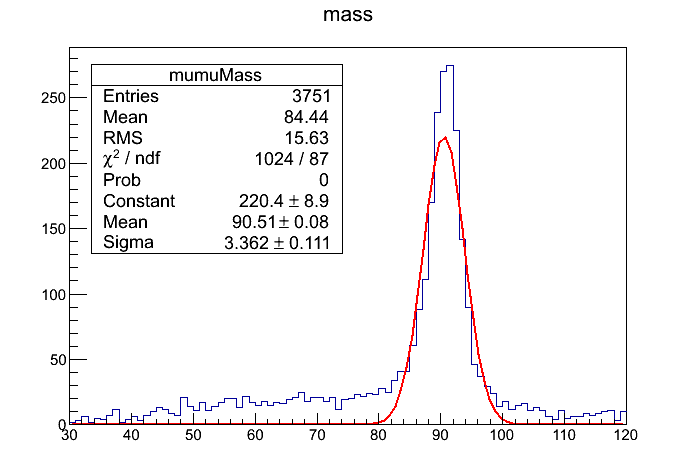
Reminder
You can quit
ROOTusing the.qcommand.
The line gStyle->SetOptFit(111111);` enables all the histogram statistics to be displayed. For more options and other information please refer to ROOT documentation.
Question 17.1a
What is the value of the mean Z Mass that you get?
Question 17.1b
What is the value of the chisquare/ndf that you get?
Using a macro of your own in ROOT
As you have seen above, we should fit a sub-range of the Z mass distribution because the fit in the full range is not all that great. In this exercise, we will fit a sub-range of the mumuMass distribution but for this we will use a ROOT macro. For more complex or useful fitting functions, one has to use a macro. The macro to run is FitZPeak.C. This macro calls another macro, BW.C. Please download/save them with the corresponding names in $CMSSW_BASE/src. Note that now the myZPeakCRAB.root file is opened by executing the macro itself, in addition to fitting the Z mass peak.
To run this macro execute the following command from the $CMSSW_BASE/src directory:
root -l FitZPeak.C
This should pop up a histogram (shown below) and you will find yourself in a ROOT session.
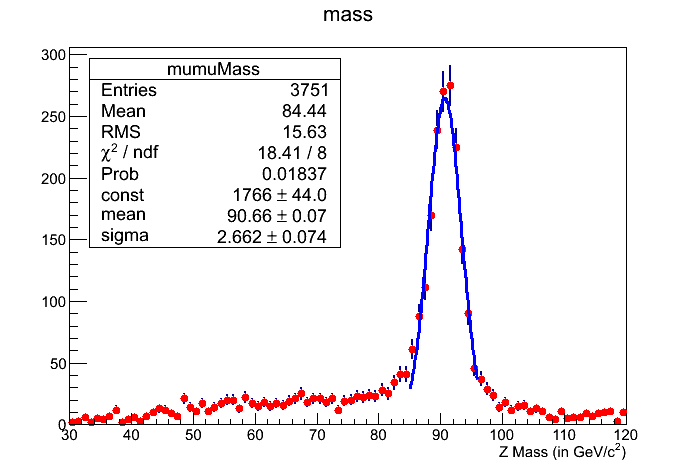
Reminder
You can save this plot from the menu on top of the histogram and then quit
ROOTusing the.qcommand.
Hint
You can also save the plot to an encapsulated postscript file by running the macro as:
root -l FitZPeak.C\(true\)
Here is some explanation of the macro. We have defined the Gaussian distribution that we want to fit in the macro BW.C (shown below). Note that in the same macro we have also is defined a Breit-Wigner function that you can try yourself. However, in the later part of the exercise, we will use RooFit to fit the distribution using a Breit-Wigner function.
Double_t mygauss(Double_t * x, Double_t * par)
{
Double_t arg = 0;
if (par[2]<0) par[2]=-par[2]; // par[2]: sigma
if (par[2] != 0) arg = (x[0] - par[1])/par[2]; // par[1]: mean
//return par[0]*BIN_SIZE*TMath::Exp(-0.5*arg*arg)/
// (TMath::Sqrt(2*TMath::Pi())*par[2]);
return par[0]*TMath::Exp(-0.5*arg*arg)/
(TMath::Sqrt(2*TMath::Pi())*par[2]); // par[0] is constant
}
par[0], par[1], and par[2] are the constant, mean, and sigma parameters, respectively. Also x[0] mean the x-axis variable. BW.C is called by FitZPeak.C in the line gROOT->LoadMacro("BW.C");. The initial values of the three fitted parameters are defined in FitZPeak.C as follows:
func->SetParameter(0,1.0); func->SetParName(0,"const");
func->SetParameter(2,5.0); func->SetParName(2,"sigma");
func->SetParameter(1,95.0); func->SetParName(1,"mean");
Also note that in the macro FitZPeak.C, we have commented the following lines and used the two lines below it. The reason being that we want to fit a sub-range. If you would want to fit the entire range of the histogram, get the minimum and maximum value of the range by instead using the lines that have been commented.
//float massMIN = Z_mass->GetBinLowEdge(1);
//float massMAX = Z_mass->GetBinLowEdge(Z_mass->GetNbinsX()+1);
float massMIN = 85.0;
float massMAX = 96.0;
Question 17.2
What mean value of the Z mass do you get in the fitted sub-range?
Using a macro in RooFit
Before we start, have a look at the RooFit twiki to get a feeling for it. Then save the macro RooFitMacro.C in the $CMSSW_BASE/src/ directory. This macro will fit the Z mass peak using RooFit.
Take a look at the code and then execute the following:
root -l RooFitMacro.C
You may need to add the following line to your rootlogon.C file to get this interpreted code to work:
gROOT->ProcessLine(".include $ROOFITSYS/include/");
This should pop a histogram (shown below) and you will find yourself in a ROOT session.
Reminder
You can save this plot from the menu on top of the histogram and then quit
ROOTusing the.qcommand.
We fit the distribution with a Gaussian by default. However, we can fit a Breit-Wigner or Voigtian (convolution of Breit-Wigner and Gaussian) by uncommenting the appropriate lines.
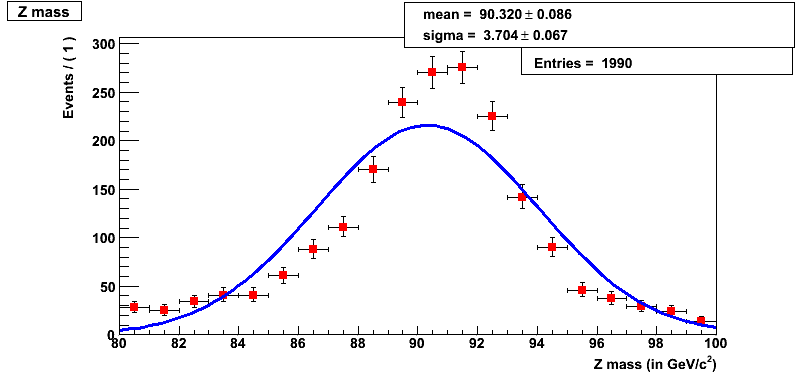
Question 17.3a
What is the mean for the gaussian fit in RooFit?
Question 17.3b
What is the sigma for the gaussian fit in RooFit?
Fitting a Breit-Wigner
Using a macro in ROOT
To fit the Z mass peak using a Breit-Wigner distribution, we first uncomment the Breit-Wigner part of FitZPeak.C and comment out the Gaussian part as follows (using /* and */):
////////////////
//For Gaussian//
///////////////
/*
TF1 *func = new TF1("mygauss",mygauss,massMIN, massMAX,3);
func->SetParameter(0,1.0); func->SetParName(0,"const");
func->SetParameter(2,5.0); func->SetParName(2,"sigma");
func->SetParameter(1,95.0); func->SetParName(1,"mean");
Z_mass->Fit("mygauss","QR");
TF1 *fit = Z_mass->GetFunction("mygauss");
*/
/////////////////////
// For Breit-Wigner//
////////////////////
TF1 *func = new TF1("mybw",mybw,massMIN, massMAX,3);
func->SetParameter(0,1.0); func->SetParName(0,"const");
func->SetParameter(2,5.0); func->SetParName(1,"sigma");
func->SetParameter(1,95.0); func->SetParName(2,"mean");
Z_mass->Fit("mybw","QR");
TF1 *fit = Z_mass->GetFunction("mybw");
Then execute the following:
root -l FitZPeak.C
This should pop a histogram (shown below) and you will find yourself in ROOT seession.
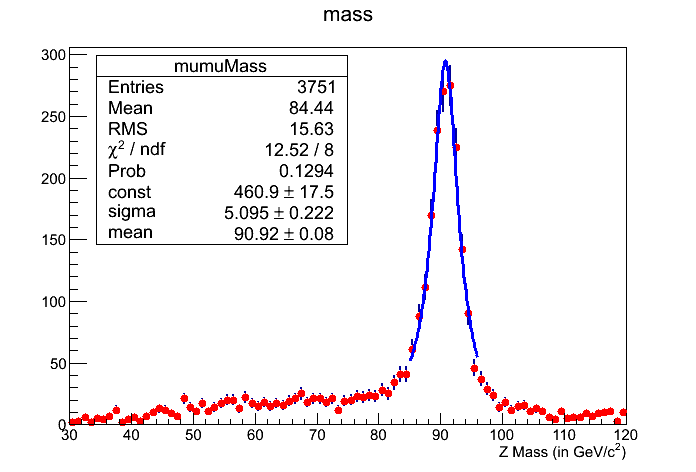
Reminder
You can save this plot from the menu on top of the histogram and then quit
ROOTusing the.qcommand.
Question 17.4a
What is the mean for the Breit-Wigner fit using the macro?
Question 17.4b
What is the sigma for Breit-Wigner fit using the macro?
Using a macro in RooFit
Before we proceed we need to uncomment and comment out few lines in RooFitMacro.C to have them look as follows:
//RooGaussian gauss("gauss","gauss",x,mean,sigma);
RooBreitWigner gauss("gauss","gauss",x,mean,sigma);
// RooVoigtian gauss("gauss","gauss",x,mean,width,sigma);
Then execute:
root -l RooFitMacro.C
This should pop a histogram (shown below) as follows and you will find yourself in ROOT session.
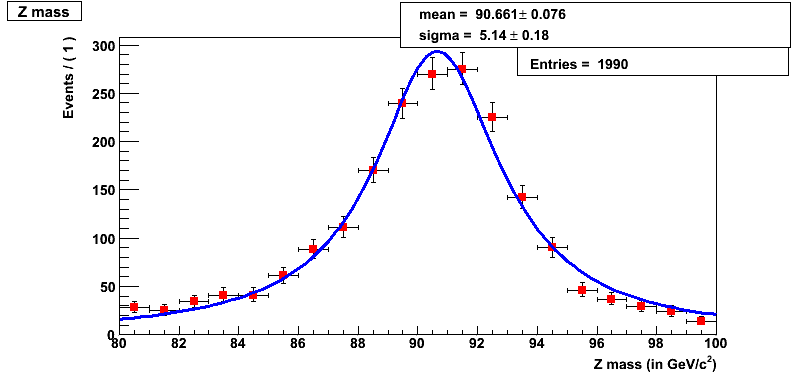
Reminder
You can save this plot from the menu on top of the histogram and then quit
ROOTusing the.qcommand.
Question 17.5a
What is the mean for the Breit-Wigner fit using RooFit tool?
Question 17.5b
What is the sigma for the Breit-Wigner fit using RooFit tool?
Fitting a Convolution of Gaussian and Breit-Wigner
Using a macro in RooFit
Before we proceed we need to uncomment and comment out few lines in RooFitMacro.C to have them look as follows:
//RooGaussian gauss("gauss","gauss",x,mean,sigma);
// RooBreitWigner gauss("gauss","gauss",x,mean,sigma);
RooVoigtian gauss("gauss","gauss",x,mean,width,sigma);
Then execute:
root -l RooFitMacro.C
This should pop a histogram (shown below) as follows and you will find yourself in ROOT seession.
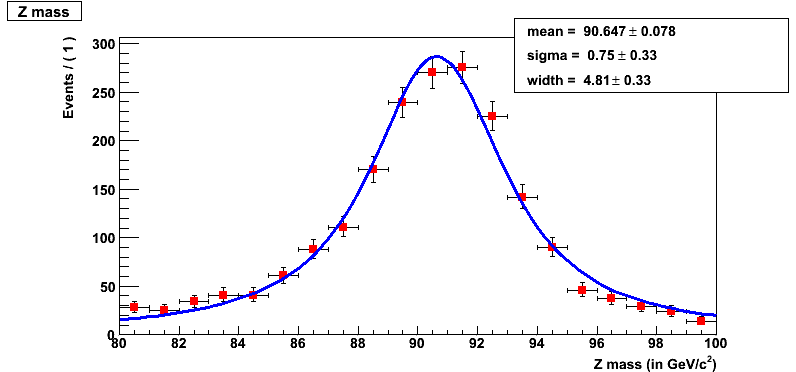
Reminder
You can save this plot from the menu on top of the histogram and then quit
ROOTusing the.qcommand.
Question 17.6a
What is the mean for the convolved fit using RooFit tool?
Question 17.6b
What is the sigma for the convolved fit using RooFit tool?
Key Points
You can use both an EDAnalyzer or FWLite to analyze MiniAOD files
Various methods exist for performing fits. You can use inbuilt functions or user defined functions. You can use plain ROOT or the RooFit package.